Introduction¶
What is protlearn?¶
protlearn is a feature extraction tool for protein sequences. It is a freely available Python package that allows the user to efficiently extract amino acid sequence features from proteins or peptides, which can then be used for a variety of downstream machine learning tasks.
Package Contents¶
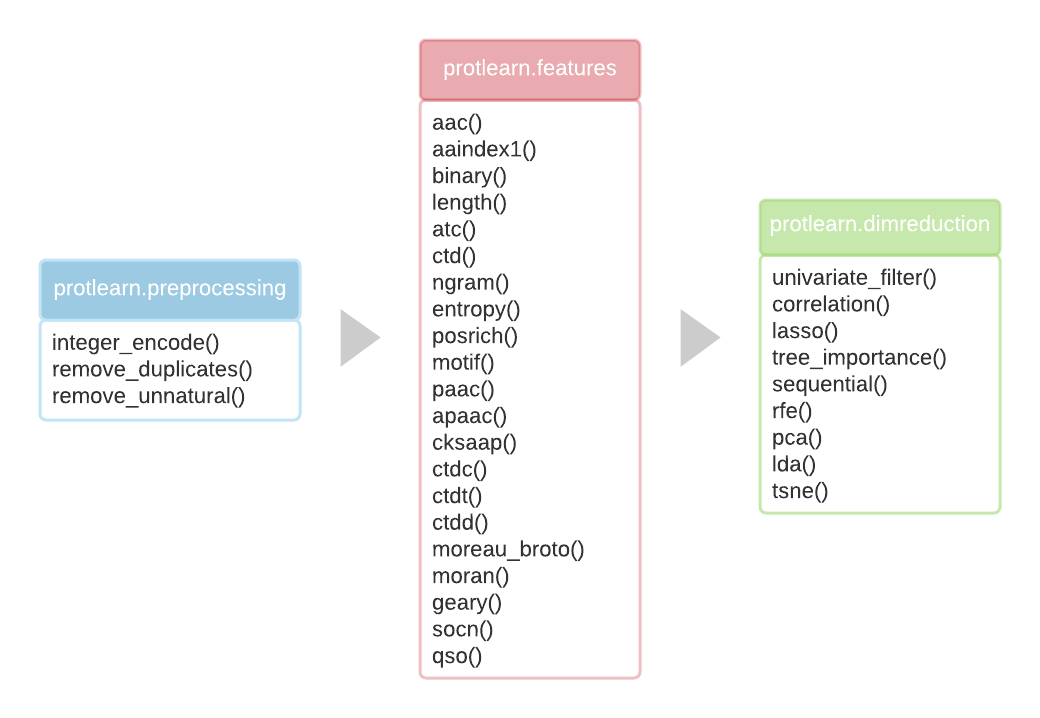
protlearn is currenty comprised of three main modules:
- preprocessing
- feature extraction
- dimensionality reduction
The Preprocessing section allows the user to prepare the datasets according to specific needs, such as filtering out sequences containing unnatural amino acids or converting the alphabetical strings to integers. The Feature Extraction section can then be used to compute amino acid sequence features from the dataset, such as amino acid composition or AAIndex-based physicochemical properties. In total, protlearn currently provides 21 such features. Finally, Dimensionality Reduction & Feature Selection methods are provided to reduce the dimensionality of the computed features, which reduces redundancy and alleviates the computational burden for the classifiers.
Input Formats¶
protlearn was designed to handle both single and multiple sequence inputs in the following formats:
- string (single)
- .fasta (single)
- list of strings (multiple)
- .fasta (multiple)